Embedded Embeddings Database: Building a low cost serverless RAG solution
The problem
Retrieval-Augmented Generation (RAG) solutions are an impressive way to talk to one’s data. One of the challenges of RAG solutions is the associated cost, often driven by the vector database. In a previous blog article I presented how to tackle this issue by using Athena with Locality Sensitive Hashing (LSH) as a knowledge database. One the of the main limitations with Athena is the latency and the low number of concurrent queries. In this new blog article, I present a new low-cost serverless solution that makes use of an embedded vector database, SQLite, to achieve a low cost while maintaining high concurrency.
Embedding databases
The following diagram presents the solution architecture.
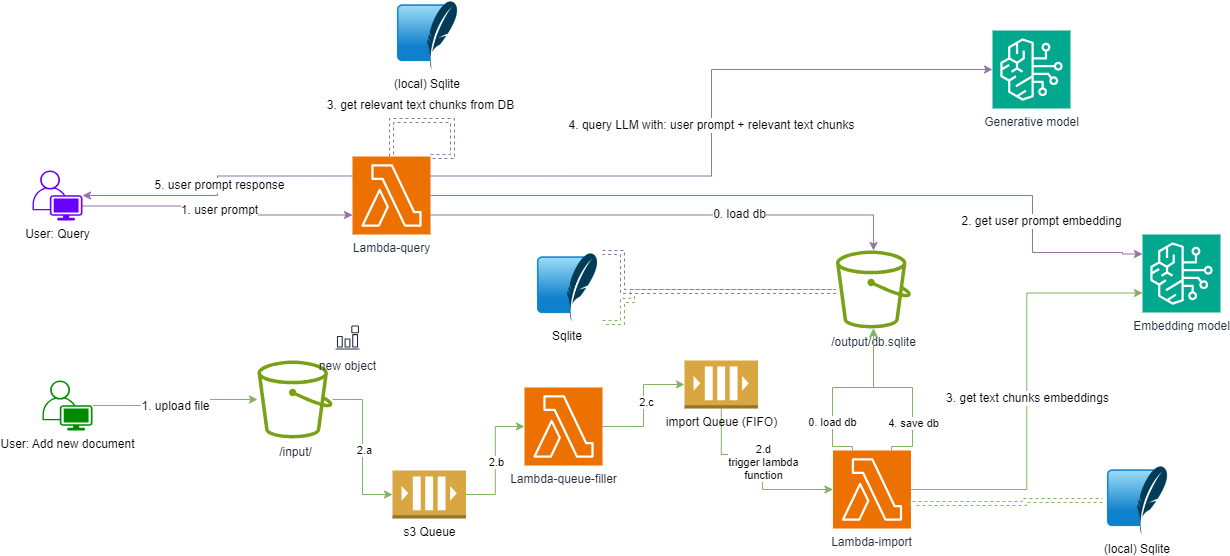 The solution uses a SQLite database that resides in an S3 bucket at rest. For queries and imports, the SQLite database is loaded into the corresponding Lambda function. The import Lambda function ensures that all changes are uploaded back to the S3 bucket.
The solution uses a SQLite database that resides in an S3 bucket at rest. For queries and imports, the SQLite database is loaded into the corresponding Lambda function. The import Lambda function ensures that all changes are uploaded back to the S3 bucket.
At the infrastructure level, we need to ensure that the Lambda-import function is never invoked concurrently, as this could otherwise result in data loss. This can been seen as an anti-pattern, and AWS currently does not allow this for a lambda function that is triggered by an SQS Queue. As a workaround we use two queues:
- s3 Queue: A queue that collects all object-creation related events, or in our case, all created file events, so that the files can be later added to the RAG knowledge database.
- import Queue: A FIFO queue with a single group ID that will trigger the Lambda-import function. Having a single group ID for all messages in the queue gives us the guarantee that the queue will never trigger multiple instances of the lambda function.
Delayed database updates: The Lambda-query function only downloads the sqlite database during cold starts. This means that even if the database is a few hundred MBs, warm starts aren’t affected at all by the download time of the database file, allowing the Lambda function to query the vector database at a blazing fast speed. However, this also means the local copy of the database is only synced during cold starts. In case of intensive usage, it can take up to a few hours for all active lambda instances to be stopped or restarted. Therefore, the Lambda-query function might, in some edge cases, need a few hours to have the updated version of the database locally. This isn’t much of an issue as imports for RAG solutions aren’t generally done in a near real-time fashion.
Getting the right Lambda environment
The whole solution relies on the sqlite-vss extension, which adds vector search functionalities to SQLite. To use it, one needs to install the extension with the corresponding Python package and load the extension into the SQLite database.
The first challenge when trying to load the extension in a Lambda function is that the Python runtime available in AWS Lambda was not installed in a way that allows SQLite to load extensions, which results in the following error when trying to load an extension into SQLite:

To solve this issue, we use a custom Docker image on which we install the runtime interface client for Python. The following snippet shows the part of the Dockerfile that is relevant to our solution.
...
# ### Customization start ###
# Install sqlite-vss dependencies
RUN apt update && apt install libgomp1 libatlas3-base -y
# Copy python requirements file
COPY requirements.txt ${LAMBDA_TASK_ROOT}
# Install python requirements
RUN python3 -m pip install -r requirements.txt --target ${FUNCTION_DIR}
# Copy python code to target dir
COPY src ${FUNCTION_DIR}/
# ### Customization end ###
...
Database design and interactions
We now have a Docker-based Lambda image in which we can load SQLite extensions.
Let’s define our database tables structure. We need two tables: A main table and an embeddings table. To the question: Why don’t we simply use a single table ? Let’s say it’s an sqlite-vss restriction.
-- main table
CREATE TABLE IF NOT EXISTS documents (
id INTEGER PRIMARY KEY AUTOINCREMENT,
text TEXT,
timestamp DATETIME
);
-- embeddings table
CREATE virtual table IF NOT EXISTS vss_documents using vss0(
text_embedding({embedding_size})
);
Before doing anything with the database, we have to make sure that the sqlite-vss extension is loaded into database. We use the following code snippet to initialize the database by loading the sqlite-vss extension and creating all needed tables.
import sqlite3
import sqlite_vss
...
def initialize_db(database: str | bytes = ON_DISK_DATABASE, embedding_size: int = EMBEDDING_SIZE):
db: sqlite3.Connection = sqlite3.connect(database)
db.enable_load_extension(True)
sqlite_vss.load(db)
db.enable_load_extension(False)
db.execute(SQL_CREATE_DOCUMENTS_TABLE)
db.execute(SQL_CREATE_VSS_DOCUMENTS_TABLE_TEMPLATE.format(embedding_size=embedding_size))
return db
To query similar embeddings from the database, we use the following SQL statement:
-- SQL_QUERY_TEMPLATE_STARTING
WITH matched_documents (rowid, distance) AS (
SELECT
rowid,
distance
FROM vss_documents
WHERE
vss_search(
text_embedding,
vss_search_params(
?, -- embedding
{top_n_documents}
)
)
ORDER BY distance ASC
)
SELECT
d.*,
m.distance
FROM matched_documents m
LEFT JOIN documents d ON m.rowid=d.rowid
WHERE
m.distance <= {distance_threshold}
ORDER BY m.distance ASC
The temporary table matched_documents contains ids and distances of matching documents. We use it to find the actual documents from the documents table, as we need the text from each document.
We are limited in the way we can query the embeddings table in SQLite versions prior to 3.41.0. Due to a bug in prior versions of SQLite, a limit statement can not be used on the embeddings table as it is a virtual table, so the number of results has to be limited by using vss_search_params, as shown in the previous SQL snippet.
When trying to query the embeddings before having saved any documents into the embeddings table, you may encounter the following error from the sqlite-vss extension, depending on your SQLite version:

The error seems to occur with SQLite versions prior to 3.41.0. To be on the safe side, we avoid querying the knowledge database when it is empty. We use the following Python function to query documents given an embedding:
def query_db_documents(
embedding: list[float] | np.ndarray,
connection: sqlite3.Connection,
top_n_documents: int = TOP_N_DOCUMENTS,
distance_threshold: float = MAX_DISTANCE_THRESHOLD,
) -> list[dict]:
serialized_embedding = get_serialized_embedding(embedding)
SQL_QUERY_VSS_DOCUMENTS_COUNT = """SELECT count(*) FROM vss_documents"""
query = SQL_QUERY_DOCUMENTS_TEMPLATE.format(top_n_documents=top_n_documents, distance_threshold=distance_threshold)
cursor: sqlite3.Cursor = connection.cursor()
cursor.row_factory = sqlite3.Row
vss_documents_count: int = cursor.execute(SQL_QUERY_VSS_DOCUMENTS_COUNT).fetchone()[0]
result: list[dict] = []
if vss_documents_count > 0:
result_rows = cursor.execute(query, [serialized_embedding])
result = [dict(item) for item in result_rows]
return result
We obviously need to be able to add documents to our knowledge database. The SQL snippets for inserts are quite straightforward:
-- SQL_INSERT_DOCUMENT
INSERT INTO documents(text, timestamp)
VALUES(:text, :timestamp)
-- SQL_INSERT_VSS_DOCUMENT
INSERT INTO vss_documents(rowid, text_embedding)
VALUES(:rowid, :text_embedding)
We use them as-is with the following Python function to save new documents into the knowledge database.
def save_document_into_db(
document: dict,
connection: sqlite3.Connection,
sql_insert_document_query: str = SQL_INSERT_DOCUMENT,
sql_insert_vss_document_query: str = SQL_INSERT_VSS_DOCUMENT,
):
document_clone = document.copy()
if "timestamp" not in document_clone:
document_clone["timestamp"] = datetime.datetime.now()
embedding: list[float] | np.ndarray = document_clone.pop("embedding")
serialized_embedding = get_serialized_embedding(embedding)
document_lastrowid = connection.execute(sql_insert_document_query, document_clone).lastrowid
vss_document_lastrowid = connection.execute(
sql_insert_vss_document_query, [document_lastrowid, serialized_embedding]
).lastrowid
return document_lastrowid == vss_document_lastrowid
The documents table and the embeddings table share the same rowid values. This is another limitation of SQLite/sqlite-vss.
So far, we have assumed that the SQLite database was available locally. However, this is only the case during warm starts of our Lambda functions. During cold starts, the database has to be downloaded from the S3 bucket and saved locally. We do it with the following python code:
def get_s3_file_locally(bucket: str, key: str, local_path: str = DEFAULT_LOCAL_DB_PATH):
try:
s3_client.head_object(Bucket=bucket, Key=key)
with open(local_path, "wb") as file_out:
s3_client.download_fileobj(bucket, key, file_out)
except botocore.exceptions.ClientError as err:
if err.response["Error"]["Code"] == "404":
# The file does not exists yet, so we will just create a new one locally
print("File not existing yet, but it is fine!")
pass
else:
raise
return local_path
It is totally fine if the database does not exists, as it will be created locally by the initialization function.
Lambda handlers
The lambda-query function isn’t much different that the one used for the Athena based RAG solution so we will only have a look at the Lambda-import function.
The lambda_handler is invoked when the sqlite database is locally available. As previously mentioned, the download from S3 only happens during cold-starts. The lambda_handler processes all elements and collect items that couldn’t be processed to report them. The s3 record processing function loads and clean the file, convert it into database items with the corresponding embeddings and save the documents into the database.
def lambda_handler(event: dict[str, object], context: dict[str, object]):
sqs_batch_response: dict = {"batch_item_failures": []}
silenced_errors = []
if event:
records = event["Records"]
batch_item_failures = []
for record in records:
record_body_reconstructed: dict[str, str | list | dict] = json.loads(record["body"])
try:
sqs_records: list[dict] = record_body_reconstructed["Records"]
for sqs_record in sqs_records:
s3_records: list[dict] = json.loads(sqs_record["body"])["Records"]
for s3_record in s3_records:
process_single_s3_record(s3_record)
except Exception as e:
batch_item_failures.append({"itemIdentifier": record["messageId"]})
silenced_errors.append(str(e))
sqs_batch_response["batchItemFailures"] = batch_item_failures
upload_file(SQLITE_DB_S3_BUCKET, SQLITE_DB_S3_KEY)
if silenced_errors:
print("SILENCED ERRORS: ", silenced_errors)
return sqs_batch_response
At the end of the Lambda-import function, the local SQLite database is uploaded to the S3 bucket. Since the Lambda-import function is not triggered concurrently and is the only one to update the database file on S3, we are guaranteed that a running instance of this lambda function always has the latest version of the database and can therefore overwrite the version in S3 without any issues.
The full code is available in the companion repository on Github.
When deployed, the stack outputs the S3 URI where documents can be uploaded to be added to the knowledge database, along with some other helpful information.
Outputs:
LowCostServerlessRAGStack.DocumentImportFolder = s3://lowcostserverlessragsqlitestack-rag70e8-pr5zms/input/
LowCostServerlessRAGSQLiteStack.LambdaQueryName = LowCostServerlessRAGSQLit-RAGDocumentQuery415C904-4kzeQQkSzUOk
Conclusion
In this blog article, we discussed how it is possible to get away from classical knowledge databases while still having a highly concurrent, low-cost solution. The presented solution comes with a few limitations: The overall database size can not be more than 10GB, a single columns can not contain more than 1GB of data and cold starts of the lambda functions grows with the database size. If most queries hit cold starts, it may make be helpful to have the database integrated into the Docker container and updated daily to further reduce costs instead of having it downloaded from S3. Nevertheless, this solution provides some nice advantages, such as being able to scale as much as the computing solution allows, and almost no idle costs, which makes it possible to have a highly scalable solution that does not need to be shut down when not in use.
The author of SQLite-VSS is now working on an extremely small successor to sqlite-vss that should work everywhere SQLite works, and without any extras: SQLite-vec. You may want to have a look at it.
— Franck
Links:
